Code
library(ggplot2)This page has code, images, and data visualizations that support the material presented in the workshop.
To reveal or hide the code, click on the button at the upper right.
Code hiding (folding) and showing is controlled by the code-fold and code-tools parameters in the YAML header for your document. See resources.qmd document to see how this is done.
We try to practice what we preach, so we include below the code we use to download the registration data and visualize it. Click on the code arrow to show or hide the code. The data file itself remains private. We have added csv/ to the .gitignore file so that the data remains available only to users who have it locally on their computers.
Rick Gilmore has stored his Gmail account information in an environment variable called GMAIL_SURVEY.
To do something similar, use the usethis package to open your .Renviron variable.
install.packages("usethis") # if not installed already
usethis::edit_r_environ()Add an entry like GMAIL_SURVEY="your.google.acct@gmail.com" to the file. Save it, and restart R.
Then, run the following line in your console:
install.packages("googledrive") # if not installed already
googledrive::drive_auth()You will see something like this:

Select ‘1’ to initiate a Google authentication screen in your default browser. The browser should launch with a window asking your permission to authorize access to your Google account. Grant access to Tidyverse API Packages. Remember, you continue to control what code from the Tidyverse accesses specific Google sheets.
Once these steps are complete, the automated updating code below should work.
We download the data as a CSV (if params$update_data == TRUE).
update_figs = TRUE
update_date_time = Sys.Date()
# Create include/csv if it doesn't exist
csv_path <- file.path(here::here(), "src", "include", "csv")
if (!dir.exists(csv_path)) {
message("Creating missing '", csv_path, "'.")
dir.create(csv_path)
}
if (params$update_data) {
options(gargle_oauth_email = Sys.getenv("GMAIL_SURVEY"))
googledrive::drive_auth()
csv_full_fn <- file.path(csv_path, params$csv_fn)
# If no file (1st run of script), create
if (!file.exists(csv_full_fn)) {
x <- data.frame(x="")
readr::write_csv(x, csv_full_fn)
}
# new_fn <- csv_full_fn
# file.rename(new_fn, stringr::str_replace(new_fn, "data-", "_data-"))
googledrive::drive_download(
params$google_fn,
path = csv_full_fn,
type = "csv",
overwrite = TRUE
)
message("Data updated.")
} else {
if (!file.exists(new_fn)) {
warning("File not found: '", new_fn, "'.")
update_figs = FALSE
} else {
message("Using previously stored data.")
}
}We load the saved CSV.
Then we clean the data by renaming the variables, dropping the “no’s”, wrangling the unit names, and adding a college variable.
TO-DO: Remove duplicate entries. Drop timestamp beforehand since it will be unique even among duplicates in other fields.
registrations_clean <- registrations |>
dplyr::rename(
timestamp = "Timestamp",
email = 'Email Address',
attend = "Will you attend the workshop on Thursday, March 27, 2025?",
name = "What is your name?",
unit = "What is your department or unit?",
position = "What is your current position?",
comments = "Any comments?"
) |>
# Remove duplicate rows
dplyr::distinct() |>
# Convert timestamp into proper dates
dplyr::mutate(timestamp = lubridate::mdy_hms(timestamp,
tz = "America/New_York")) |>
# Normalize unit names
dplyr::mutate(
unit = dplyr::recode(
unit,
`ESM-Ecosystem Science and Mgmt` = "Ecosystem Science & Mgmt",
`Earth and Environmental Systems Institute` = "Earth & Environmental Systems Institute",
`Earth and Environmental Systems Institute` = "Earth & Environmental Systems Institute",
`Civil and Environmental Engineering` = "Civil & Environmental Engineering",
`Civil and Environmental Eng` = "Civil & Environmental Engineering",
`chemical engineering` = "Chemical Engineering",
`FOOD SCIENCE` = "Food Science",
`Earth and Mineral Sciences, Energy Institute` = "Energy Institute",
`Center for Healthy Aging / HHD` = "Center for Healthy Aging",
`BMB` = "Biochemistry, Microbiology, & Molecular Biology",
`Biochemistry, Microbiology and Molecular Biology` = "Biochemistry, Microbiology, & Molecular Biology",
`BMMB` = "Biochemistry, Microbiology, & Molecular Biology",
`Spanish, Italian and Portuguese` = "Spanish, Italian, & Portuguese",
`Spanish, Italian, and Portuguese` = "Spanish, Italian, & Portuguese",
`SIP` = "Spanish, Italian, & Portuguese",
`Department of Nutritional Sciences` = "Nutritional Sciences",
`Department of Medicine, Division of Endocrinology` = "Medicine",
`Nutrition` = "Nutritional Sciences",
`Department of Statistics` = "Statistics",
`Dept of Statistics` = "Statistics",
`RPTM` = "Recreation, Park, & Tourism Management",
`Kinesiology & Developmental Psychology` = "Kinesiology",
`Educational psychology` = "Educational Psychology",
`Clinical Psychology` = "Psychology",
`Developmental Psychology` = "Psychology",
`Communication Sciences and Disorders` = "Communication Sciences & Disorders",
`Center for Healthy Aging; Department of Psychology` = "Center for Healthy Aging",
`MCIBS` = "Molecular, Cellular, & Integrative Biosciences",
`Department of Chemical Engineering` = "Chemical Engineering",
`Biobehavioral health` = "Biobehavioral Health",
`EE` = "Electrical Engineering",
`Agricultural and biological engineering` = "Agricultural & Biological Engineering",
`Nutrition: Kathleen Keller` = "Nutritional Sciences"
)
) |>
# Drop no attends
dplyr::filter(attend == "Yes") |>
# Add college
dplyr::mutate(
college = dplyr::case_match(
unit,
"Statistics" ~ "ECoS",
"Biology" ~ "ECoS",
"Biochemistry, Microbiology, & Molecular Biology" ~ "ECoS",
"Biomedical engineering" ~ "Eng",
"Physics" ~ "ECoS",
"Chemistry" ~ "ECoS",
"Astronomy & Astrophysics" ~ "ECoS",
"Eberly" ~ "ECos",
"Eberly" ~ "ECos",
"Eberly College of Science" ~ "ECoS",
"ECos" ~ "ECoS",
"Poli Sci" ~ "CLA",
"Prevention research center" ~ "HHD",
"Center for Healthy Aging (HHD)" ~ "HHD",
"Sociology and Criminology" ~ "CLA",
"Psychology" ~ "CLA",
"Spanish, Italian, & Portuguese" ~ "CLA",
"Research Informatics and Publishing" ~ "Libraries",
"Political Science" ~ "CLA",
"Applied Linguistics" ~ "CLA",
"Global Languages & Literatures" ~ "CLA",
"Sociology" ~ "CLA",
"English" ~ "CLA",
"C-SoDA" ~ "CLA",
"Office of Digital Pedagogies and Initiatives" ~ "CLA",
"Asian Studies" ~ "CLA",
"Anthropology" ~ "CLA",
"Linguistics" ~ "CLA",
"Center for Language Science" ~ "CLA",
"Foreign Languages" ~ "CLA",
"Languages and Literature" ~ "CLA",
"Developmental Psychology" ~ "CLA",
"IST" ~ "IST",
"Chemical Engineering" ~ "Eng",
"Civil & Environmental Engineering" ~ "Eng",
"Material Science and Engineering" ~ "Eng",
"Engineering Science & Mechanics" ~ "Eng",
"Biomedical Engineering" ~ "Eng",
"Mechanical Engineering" ~ "Eng",
"Electrical Engineering" ~ "Eng",
"Chemical Engineering" ~ "Eng",
"Nutritional Sciences" ~ "HHD",
"HDFS" ~ "HHD",
"Communication Sciences & Disorders" ~ "HHD",
"Kinesiology" ~ "HHD",
"Recreation, Park, & Tourism Management" ~ "HHD",
"Prevention Center" ~ "HHD",
"Bellisario College of Communication" ~ "Comm",
"Marketing" ~ "Smeal",
"Food Science" ~ "Ag",
"Ecosystem Science & Mgmt" ~ "Ag",
"Entomology" ~ "Ag",
"Plant Pathology & Environmental Microbiology" ~ "Ag",
"Plant Science" ~ "Ag",
"Agricultural & Biological Engineering" ~ "Ag",
"Landscape Architecture" ~ "Arts & Arch",
"Neuroscience" ~ "Med",
"Medicine" ~ "Med",
"College of Human and Health Development" ~ "HHD",
"Center for Healthy Aging" ~ "HHD",
"Center for Childhood Obesity Research" ~ "HHD",
"Biobehavioral Health" ~ "HHD",
"Earth & Environmental Systems Institute" ~ "EMS",
"Geography" ~ "EMS",
"Energy Institute" ~ "EMS",
"Geosciences" ~ "EMS",
"Nursing" ~ "Nursing",
"Educational Psychology" ~ "Ed",
"Molecular, Cellular, & Integrative Biosciences" ~ "Huck",
"University Libraries" ~ "Libraries"
),
.default = "Unknown",
.missing = "Unknown"
)
# registrations_clean <- registrations_clean |>
# dplyr::mutate(
# confirmed = dplyr::case_match(
# confirmation_response,
# "Sorry, can't make it" ~ "Decline",
# "No response" ~ "No response",
# "Yes, I'll be there" ~ "Confirm",
# NA ~ "No response",
# "presenter" ~ "Confirm"),
# .default = "Unknown",
# .missing = "Unknown"
# )The following is used to modify the above code to capture new unit names, clean/normalize them, and assign them to colleges.
registrations_clean |>
dplyr::arrange(timestamp) |>
dplyr::mutate(resp_index = seq_along(timestamp)) |>
ggplot() +
aes(x = timestamp, y = resp_index, color = position) +
geom_point() +
theme(axis.text.x = element_text(angle = 90)) +
labs(x = NULL, y = 'n') +
theme(legend.position = "bottom", legend.title = element_blank())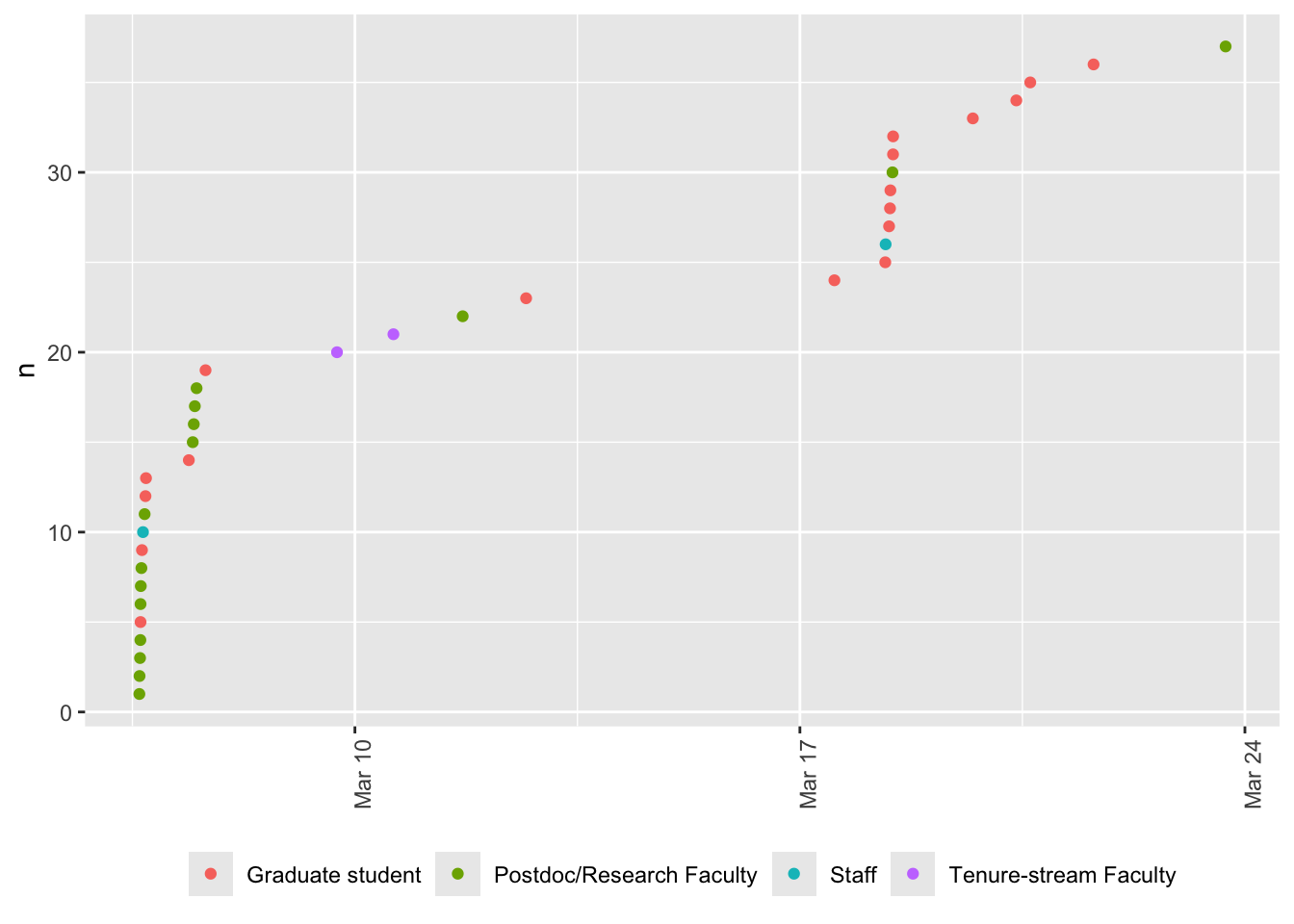
---
title: ""
format:
html:
code-fold: true
code-tools: true
toc: true
params:
update_data: true
google_fn: "Data Management Workshop Spring 2025: Registration (Responses)"
csv_fn: "data-mgmt-wksp-2025.csv"
---
## About
This page has code, images, and data visualizations that support the material presented in the workshop.
To reveal or hide the code, click on the button at the upper right.
::: {.callout-tip}
Code hiding (folding) and showing is controlled by the `code-fold` and `code-tools` parameters in the YAML header for your document.
See [`resources.qmd` document](https://github.com/penn-state-open-science/data-mgmt-2024/blob/main/src/resources.qmd) to see how this is done.
:::
## Setup
```{r}
library(ggplot2)
```
## Registrant data
We try to practice what we preach, so we include below the code we use to download the registration data and visualize it.
Click on the `code` arrow to show or hide the code.
The data file itself remains private.
We have added `csv/` to the `.gitignore` file so that the data remains available only to users who have it locally on their computers.
::: {.callout-note}
Rick Gilmore has stored his Gmail account information in an environment variable called `GMAIL_SURVEY`.
To do something similar, use the [`usethis` package](https://cran.r-project.org/package=usethis) to open your `.Renviron` variable.
```
install.packages("usethis") # if not installed already
usethis::edit_r_environ()
```
Add an entry like `GMAIL_SURVEY="your.google.acct@gmail.com"` to the file.
Save it, and restart R.
Then, run the following line in your console:
```
install.packages("googledrive") # if not installed already
googledrive::drive_auth()
```
You will see something like this:

Select '1' to initiate a Google authentication screen in your default browser.
The browser should launch with a window asking your permission to authorize access to your Google account.
Grant access to Tidyverse API Packages.
Remember, *you* continue to control what code from the Tidyverse accesses specific Google sheets.
Once these steps are complete, the *automated* updating code below should work.
:::
We download the data as a CSV (if `params$update_data == TRUE`).
```{r set-up-update-data}
#| label: gather-data-from-google
#| message: false
#| echo: true
update_figs = TRUE
update_date_time = Sys.Date()
# Create include/csv if it doesn't exist
csv_path <- file.path(here::here(), "src", "include", "csv")
if (!dir.exists(csv_path)) {
message("Creating missing '", csv_path, "'.")
dir.create(csv_path)
}
if (params$update_data) {
options(gargle_oauth_email = Sys.getenv("GMAIL_SURVEY"))
googledrive::drive_auth()
csv_full_fn <- file.path(csv_path, params$csv_fn)
# If no file (1st run of script), create
if (!file.exists(csv_full_fn)) {
x <- data.frame(x="")
readr::write_csv(x, csv_full_fn)
}
# new_fn <- csv_full_fn
# file.rename(new_fn, stringr::str_replace(new_fn, "data-", "_data-"))
googledrive::drive_download(
params$google_fn,
path = csv_full_fn,
type = "csv",
overwrite = TRUE
)
message("Data updated.")
} else {
if (!file.exists(new_fn)) {
warning("File not found: '", new_fn, "'.")
update_figs = FALSE
} else {
message("Using previously stored data.")
}
}
```
We load the saved CSV.
```{r load-registration-data}
if (update_figs) {
registrations <-
readr::read_csv(csv_full_fn, show_col_types = FALSE)
}
```
Then we clean the data by renaming the variables, dropping the "no's", wrangling the unit names, and adding a `college` variable.
::: {.callout-note}
**TO-DO:** Remove duplicate entries. Drop timestamp beforehand since it will be unique even among duplicates in other fields.
:::
```{r clean-registration-data, eval=update_figs}
registrations_clean <- registrations |>
dplyr::rename(
timestamp = "Timestamp",
email = 'Email Address',
attend = "Will you attend the workshop on Thursday, March 27, 2025?",
name = "What is your name?",
unit = "What is your department or unit?",
position = "What is your current position?",
comments = "Any comments?"
) |>
# Remove duplicate rows
dplyr::distinct() |>
# Convert timestamp into proper dates
dplyr::mutate(timestamp = lubridate::mdy_hms(timestamp,
tz = "America/New_York")) |>
# Normalize unit names
dplyr::mutate(
unit = dplyr::recode(
unit,
`ESM-Ecosystem Science and Mgmt` = "Ecosystem Science & Mgmt",
`Earth and Environmental Systems Institute` = "Earth & Environmental Systems Institute",
`Earth and Environmental Systems Institute` = "Earth & Environmental Systems Institute",
`Civil and Environmental Engineering` = "Civil & Environmental Engineering",
`Civil and Environmental Eng` = "Civil & Environmental Engineering",
`chemical engineering` = "Chemical Engineering",
`FOOD SCIENCE` = "Food Science",
`Earth and Mineral Sciences, Energy Institute` = "Energy Institute",
`Center for Healthy Aging / HHD` = "Center for Healthy Aging",
`BMB` = "Biochemistry, Microbiology, & Molecular Biology",
`Biochemistry, Microbiology and Molecular Biology` = "Biochemistry, Microbiology, & Molecular Biology",
`BMMB` = "Biochemistry, Microbiology, & Molecular Biology",
`Spanish, Italian and Portuguese` = "Spanish, Italian, & Portuguese",
`Spanish, Italian, and Portuguese` = "Spanish, Italian, & Portuguese",
`SIP` = "Spanish, Italian, & Portuguese",
`Department of Nutritional Sciences` = "Nutritional Sciences",
`Department of Medicine, Division of Endocrinology` = "Medicine",
`Nutrition` = "Nutritional Sciences",
`Department of Statistics` = "Statistics",
`Dept of Statistics` = "Statistics",
`RPTM` = "Recreation, Park, & Tourism Management",
`Kinesiology & Developmental Psychology` = "Kinesiology",
`Educational psychology` = "Educational Psychology",
`Clinical Psychology` = "Psychology",
`Developmental Psychology` = "Psychology",
`Communication Sciences and Disorders` = "Communication Sciences & Disorders",
`Center for Healthy Aging; Department of Psychology` = "Center for Healthy Aging",
`MCIBS` = "Molecular, Cellular, & Integrative Biosciences",
`Department of Chemical Engineering` = "Chemical Engineering",
`Biobehavioral health` = "Biobehavioral Health",
`EE` = "Electrical Engineering",
`Agricultural and biological engineering` = "Agricultural & Biological Engineering",
`Nutrition: Kathleen Keller` = "Nutritional Sciences"
)
) |>
# Drop no attends
dplyr::filter(attend == "Yes") |>
# Add college
dplyr::mutate(
college = dplyr::case_match(
unit,
"Statistics" ~ "ECoS",
"Biology" ~ "ECoS",
"Biochemistry, Microbiology, & Molecular Biology" ~ "ECoS",
"Biomedical engineering" ~ "Eng",
"Physics" ~ "ECoS",
"Chemistry" ~ "ECoS",
"Astronomy & Astrophysics" ~ "ECoS",
"Eberly" ~ "ECos",
"Eberly" ~ "ECos",
"Eberly College of Science" ~ "ECoS",
"ECos" ~ "ECoS",
"Poli Sci" ~ "CLA",
"Prevention research center" ~ "HHD",
"Center for Healthy Aging (HHD)" ~ "HHD",
"Sociology and Criminology" ~ "CLA",
"Psychology" ~ "CLA",
"Spanish, Italian, & Portuguese" ~ "CLA",
"Research Informatics and Publishing" ~ "Libraries",
"Political Science" ~ "CLA",
"Applied Linguistics" ~ "CLA",
"Global Languages & Literatures" ~ "CLA",
"Sociology" ~ "CLA",
"English" ~ "CLA",
"C-SoDA" ~ "CLA",
"Office of Digital Pedagogies and Initiatives" ~ "CLA",
"Asian Studies" ~ "CLA",
"Anthropology" ~ "CLA",
"Linguistics" ~ "CLA",
"Center for Language Science" ~ "CLA",
"Foreign Languages" ~ "CLA",
"Languages and Literature" ~ "CLA",
"Developmental Psychology" ~ "CLA",
"IST" ~ "IST",
"Chemical Engineering" ~ "Eng",
"Civil & Environmental Engineering" ~ "Eng",
"Material Science and Engineering" ~ "Eng",
"Engineering Science & Mechanics" ~ "Eng",
"Biomedical Engineering" ~ "Eng",
"Mechanical Engineering" ~ "Eng",
"Electrical Engineering" ~ "Eng",
"Chemical Engineering" ~ "Eng",
"Nutritional Sciences" ~ "HHD",
"HDFS" ~ "HHD",
"Communication Sciences & Disorders" ~ "HHD",
"Kinesiology" ~ "HHD",
"Recreation, Park, & Tourism Management" ~ "HHD",
"Prevention Center" ~ "HHD",
"Bellisario College of Communication" ~ "Comm",
"Marketing" ~ "Smeal",
"Food Science" ~ "Ag",
"Ecosystem Science & Mgmt" ~ "Ag",
"Entomology" ~ "Ag",
"Plant Pathology & Environmental Microbiology" ~ "Ag",
"Plant Science" ~ "Ag",
"Agricultural & Biological Engineering" ~ "Ag",
"Landscape Architecture" ~ "Arts & Arch",
"Neuroscience" ~ "Med",
"Medicine" ~ "Med",
"College of Human and Health Development" ~ "HHD",
"Center for Healthy Aging" ~ "HHD",
"Center for Childhood Obesity Research" ~ "HHD",
"Biobehavioral Health" ~ "HHD",
"Earth & Environmental Systems Institute" ~ "EMS",
"Geography" ~ "EMS",
"Energy Institute" ~ "EMS",
"Geosciences" ~ "EMS",
"Nursing" ~ "Nursing",
"Educational Psychology" ~ "Ed",
"Molecular, Cellular, & Integrative Biosciences" ~ "Huck",
"University Libraries" ~ "Libraries"
),
.default = "Unknown",
.missing = "Unknown"
)
# registrations_clean <- registrations_clean |>
# dplyr::mutate(
# confirmed = dplyr::case_match(
# confirmation_response,
# "Sorry, can't make it" ~ "Decline",
# "No response" ~ "No response",
# "Yes, I'll be there" ~ "Confirm",
# NA ~ "No response",
# "presenter" ~ "Confirm"),
# .default = "Unknown",
# .missing = "Unknown"
# )
```
The following is used to modify the above code to capture new unit names, clean/normalize them, and assign them to colleges.
```{r, eval=update_figs}
college_na <- is.na(registrations_clean$college)
registrations_clean$unit[college_na]
```
### Time series
```{r , eval=update_figs}
#| label: fig-registration-time-series
#| fig-cap: "Time series of registrations."
#| eval: update_figs
registrations_clean |>
dplyr::arrange(timestamp) |>
dplyr::mutate(resp_index = seq_along(timestamp)) |>
ggplot() +
aes(x = timestamp, y = resp_index, color = position) +
geom_point() +
theme(axis.text.x = element_text(angle = 90)) +
labs(x = NULL, y = 'n') +
theme(legend.position = "bottom", legend.title = element_blank())
```
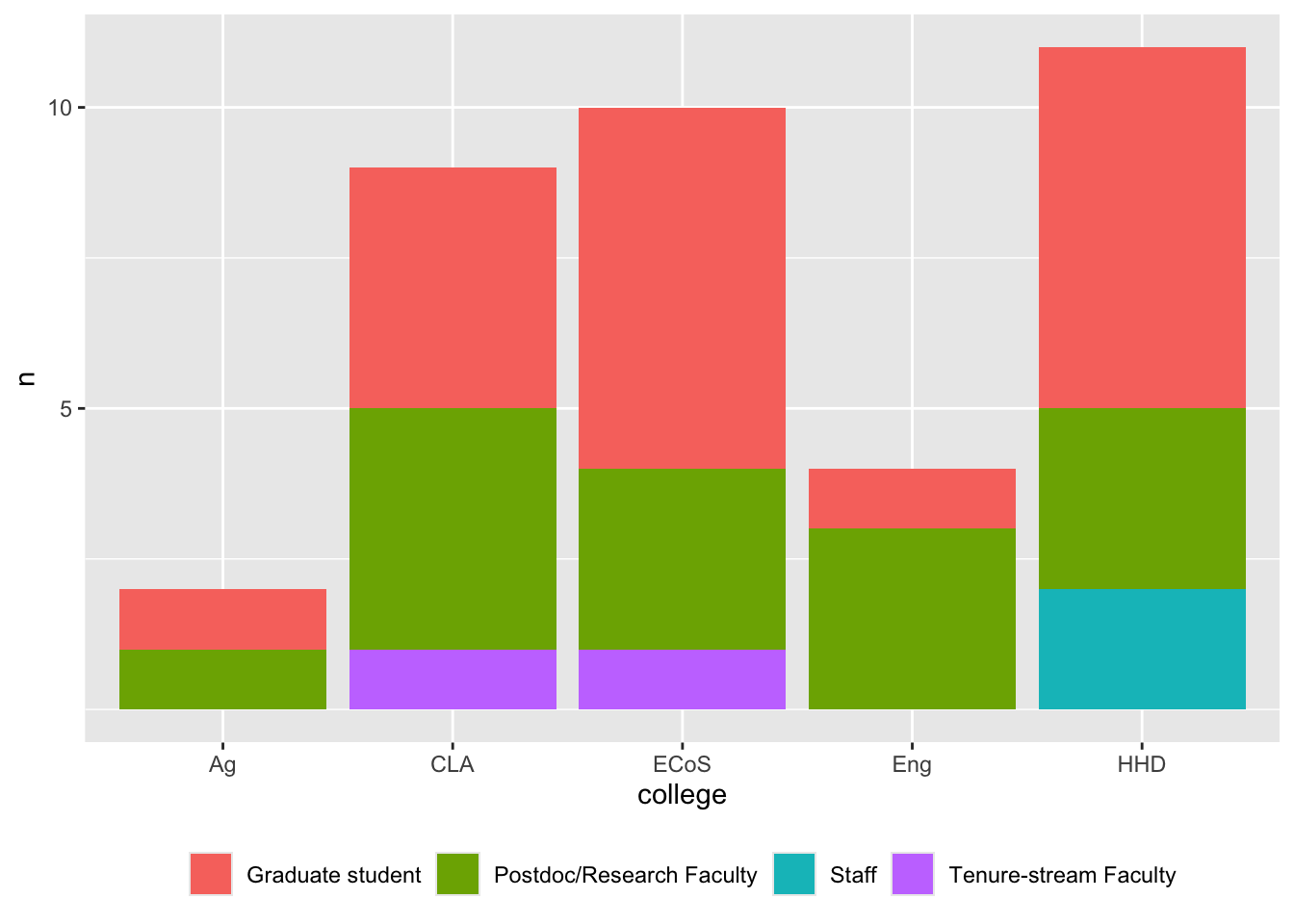
### By college and position
```{r}
#| label: fig-registrations-college-position
#| fig-cap: "Workshop registrations by college and position."
registrations_clean |>
dplyr::filter(!is.na(college)) |>
dplyr::count(position, college, sort = TRUE) |>
ggplot() +
aes(college, n, fill = position) +
geom_col() +
theme(legend.position = "bottom", legend.title = element_blank()) +
scale_y_continuous(breaks = c(5,10,15))
```